Human Tenascin X (TNX) ELISA Kit

two product lines: Traditional ELISA Kit and Ready-to-Use ELISA Kit.


Other names:TN-X; TNXB; TNX-B; TNXA; TNX-A; TNXB1; TNXB2; HXBL; Hexabrachion-like protein
Function: Appears to mediate interactions between cells and the extracellular matrix. Substrate-adhesion molecule that appears to inhibit cell migration. Accelerates collagen fibril formation. May play a role in supporting the growth of epithelial tumors.
Sequence:
10 20 30 40 50 MMPAQYALTS SLVLLVLLST ARAGPFSSRS NVTLPAPRPP PQPGGHTVGA 60 70 80 90 100 GVGSPSSQLY EHTVEGGEKQ VVFTHRINLP PSTGCGCPPG TEPPVLASEV 110 120 130 140 150 QALRVRLEIL EELVKGLKEQ CTGGCCPASA QAGTGQTDVR TLCSLHGVFD 160 170 180 190 200 LSRCTCSCEP GWGGPTCSDP TDAEIPPSSP PSASGSCPDD CNDQGRCVRG 210 220 230 240 250 RCVCFPGYTG PSCGWPSCPG DCQGRGRCVQ GVCVCRAGFS GPDCSQRSCP 260 270 280 290 300 RGCSQRGRCE GGRCVCDPGY TGDDCGMRSC PRGCSQRGRC ENGRCVCNPG 310 320 330 340 350 YTGEDCGVRS CPRGCSQRGR CKDGRCVCDP GYTGEDCGTR SCPWDCGEGG 360 370 380 390 400 RCVDGRCVCW PGYTGEDCST RTCPRDCRGR GRCEDGECIC DTGYSGDDCG 410 420 430 440 450 VRSCPGDCNQ RGRCEDGRCV CWPGYTGTDC GSRACPRDCR GRGRCENGVC 460 470 480 490 500 VCNAGYSGED CGVRSCPGDC RGRGRCESGR CMCWPGYTGR DCGTRACPGD 510 520 530 540 550 CRGRGRCVDG RCVCNPGFTG EDCGSRRCPG DCRGHGLCED GVCVCDAGYS 560 570 580 590 600 GEDCSTRSCP GGCRGRGQCL DGRCVCEDGY SGEDCGVRQC PNDCSQHGVC 610 620 630 640 650 QDGVCICWEG YVSEDCSIRT CPSNCHGRGR CEEGRCLCDP GYTGPTCATR 660 670 680 690 700 MCPADCRGRG RCVQGVCLCH VGYGGEDCGQ EEPPASACPG GCGPRELCRA 710 720 730 740 750 GQCVCVEGFR GPDCAIQTCP GDCRGRGECH DGSCVCKDGY AGEDCGEEVP 760 770 780 790 800 TIEGMRMHLL EETTVRTEWT PAPGPVDAYE IQFIPTTEGA SPPFTARVPS 810 820 830 840 850 SASAYDQRGL APGQEYQVTV RALRGTSWGL PASKTITTMI DGPQDLRVVA 860 870 880 890 900 VTPTTLELGW LRPQAEVDRF VVSYVSAGNQ RVRLEVPPEA DGTLLTDLMP 910 920 930 940 950 GVEYVVTVTA ERGRAVSYPA SVRANTGSSP LGLLGTTDEP PPSGPSTTQG 960 970 980 990 1000 AQAPLLQQRP QELGELRVLG RDETGRLRVV WTAQPDTFAY FQLRMRVPEG 1010 1020 1030 1040 1050 PGAHEEVLPG DVRQALVPPP PPGTPYELSL HGVPPGGKPS DPIIYQGIMD 1060 1070 1080 1090 1100 KDEEKPGKSS GPPRLGELTV TDRTSDSLLL RWTVPEGEFD SFVIQYKDRD 1110 1120 1130 1140 1150 GQPQVVPVEG PQRSAVITSL DPGRKYKFVL YGFVGKKRHG PLVAEAKILP 1160 1170 1180 1190 1200 QSDPSPGTPP HLGNLWVTDP TPDSLHLSWT VPEGQFDTFM VQYRDRDGRP 1210 1220 1230 1240 1250 QVVPVEGPER SFVVSSLDPD HKYRFTLFGI ANKKRYGPLT ADGTTAPERK 1260 1270 1280 1290 1300 EEPPRPEFLE QPLLGELTVT GVTPDSLRLS WTVAQGPFDS FMVQYKDAQG 1310 1320 1330 1340 1350 QPQAVPVAGD ENEVTVPGLD PDRKYKMNLY GLRGRQRVGP ESVVAKTAPQ 1360 1370 1380 1390 1400 EDVDETPSPT ELGTEAPESP EEPLLGELTV TGSSPDSLSL FWTVPQGSFD 1410 1420 1430 1440 1450 SFTVQYKDRD GRPRAVRVGG KESEVTVGGL EPGHKYKMHL YGLHEGQRVG 1460 1470 1480 1490 1500 PVSAVGVTAP QQEETPPATE SPLEPRLGEL TVTDVTPNSV GLSWTVPEGQ 1510 1520 1530 1540 1550 FDSFIVQYKD KDGQPQVVPV AADQREVTVY NLEPERKYKM NMYGLHDGQR 1560 1570 1580 1590 1600 MGPLSVVIVT APLPPAPATE ASKPPLEPRL GELTVTDITP DSVGLSWTVP 1610 1620 1630 1640 1650 EGEFDSFVVQ YKDRDGQPQV VPVAADQREV TIPDLEPSRK YKFLLFGIQD 1660 1670 1680 1690 1700 GKRRSPVSVE AKTVARGDAS PGAPPRLGEL WVTDPTPDSL RLSWTVPEGQ 1710 1720 1730 1740 1750 FDSFVVQFKD KDGPQVVPVE GHERSVTVTP LDAGRKYRFL LYGLLGKKRH 1760 1770 1780 1790 1800 GPLTADGTTE ARSAMDDTGT KRPPKPRLGE ELQVTTVTQN SVGLSWTVPE 1810 1820 1830 1840 1850 GQFDSFVVQY KDRDGQPQVV PVEGSLREVS VPGLDPAHRY KLLLYGLHHG 1860 1870 1880 1890 1900 KRVGPISAVA ITAGREETET ETTAPTPPAP EPHLGELTVE EATSHTLHLS 1910 1920 1930 1940 1950 WMVTEGEFDS FEIQYTDRDG QLQMVRIGGD RNDITLSGLE SDHRYLVTLY 1960 1970 1980 1990 2000 GFSDGKHVGP VHVEALTVPE EEKPSEPPTA TPEPPIKPRL GELTVTDATP 2010 2020 2030 2040 2050 DSLSLSWTVP EGQFDHFLVQ YRNGDGQPKA VRVPGHEEGV TISGLEPDHK 2060 2070 2080 2090 2100 YKMNLYGFHG GQRMGPVSVV GVTAAEEETP SPTEPSMEAP EPAEEPLLGE 2110 2120 2130 2140 2150 LTVTGSSPDS LSLSWTVPQG RFDSFTVQYK DRDGRPQVVR VGGEESEVTV 2160 2170 2180 2190 2200 GGLEPGRKYK MHLYGLHEGR RVGPVSAVGV TAPEEESPDA PLAKLRLGQM 2210 2220 2230 2240 2250 TVRDITSDSL SLSWTVPEGQ FDHFLVQFKN GDGQPKAVRV PGHEDGVTIS 2260 2270 2280 2290 2300 GLEPDHKYKM NLYGFHGGQR VGPVSAVGLT APGKDEEMAP ASTEPPTPEP 2310 2320 2330 2340 2350 PIKPRLEELT VTDATPDSLS LSWTVPEGQF DHFLVQYKNG DGQPKATRVP 2360 2370 2380 2390 2400 GHEDRVTISG LEPDNKYKMN LYGFHGGQRV GPVSAIGVTA AEEETPSPTE 2410 2420 2430 2440 2450 PSMEAPEPPE EPLLGELTVT GSSPDSLSLS WTVPQGRFDS FTVQYKDRDG 2460 2470 2480 2490 2500 RPQVVRVGGE ESEVTVGGLE PGRKYKMHLY GLHEGRRVGP VSTVGVTAPQ 2510 2520 2530 2540 2550 EDVDETPSPT EPGTEAPGPP EEPLLGELTV TGSSPDSLSL SWTVPQGRFD 2560 2570 2580 2590 2600 SFTVQYKDRD GRPQAVRVGG QESKVTVRGL EPGRKYKMHL YGLHEGRRLG 2610 2620 2630 2640 2650 PVSAVGVTED EAETTQAVPT MTPEPPIKPR LGELTMTDAT PDSLSLSWTV 2660 2670 2680 2690 2700 PEGQFDHFLV QYRNGDGQPK AVRVPGHEDG VTISGLEPDH KYKMNLYGFH 2710 2720 2730 2740 2750 GGQRVGPISV IGVTAAEEET PSPTELSTEA PEPPEEPLLG ELTVTGSSPD 2760 2770 2780 2790 2800 SLSLSWTIPQ GHFDSFTVQY KDRDGRPQVM RVRGEESEVT VGGLEPGRKY 2810 2820 2830 2840 2850 KMHLYGLHEG RRVGPVSTVG VTAPEDEAET TQAVPTTTPE PPNKPRLGEL 2860 2870 2880 2890 2900 TVTDATPDSL SLSWMVPEGQ FDHFLVQYRN GDGQPKVVRV PGHEDGVTIS 2910 2920 2930 2940 2950 GLEPDHKYKM NLYGFHGGQR VGPISVIGVT AAEEETPAPT EPSTEAPEPP 2960 2970 2980 2990 3000 EEPLLGELTV TGSSPDSLSL SWTIPQGRFD SFTVQYKDRD GRPQVVRVRG 3010 3020 3030 3040 3050 EESEVTVGGL EPGCKYKMHL YGLHEGQRVG PVSAVGVTAP KDEAETTQAV 3060 3070 3080 3090 3100 PTMTPEPPIK PRLGELTVTD ATPDSLSLSW MVPEGQFDHF LVQYRNGDGQ 3110 3120 3130 3140 3150 PKAVRVPGHE DGVTISGLEP DHKYKMNLYG FHGGQRVGPV SAIGVTEEET 3160 3170 3180 3190 3200 PSPTEPSTEA PEAPEEPLLG ELTVTGSSPD SLSLSWTVPQ GRFDSFTVQY 3210 3220 3230 3240 3250 KDRDGQPQVV RVRGEESEVT VGGLEPGRKY KMHLYGLHEG QRVGPVSTVG 3260 3270 3280 3290 3300 ITAPLPTPLP VEPRLGELAV AAVTSDSVGL SWTVAQGPFD SFLVQYRDAQ 3310 3320 3330 3340 3350 GQPQAVPVSG DLRAVAVSGL DPARKYKFLL FGLQNGKRHG PVPVEARTAP 3360 3370 3380 3390 3400 DTKPSPRLGE LTVTDATPDS VGLSWTVPEG EFDSFVVQYK DKDGRLQVVP 3410 3420 3430 3440 3450 VAANQREVTV QGLEPSRKYR FLLYGLSGRK RLGPISADST TAPLEKELPP 3460 3470 3480 3490 3500 HLGELTVAEE TSSSLRLSWT VAQGPFDSFV VQYRDTDGQP RAVPVAADQR 3510 3520 3530 3540 3550 TVTVEDLEPG KKYKFLLYGL LGGKRLGPVS ALGMTAPEED TPAPELAPEA 3560 3570 3580 3590 3600 PEPPEEPRLG VLTVTDTTPD SMRLSWSVAQ GPFDSFVVQY EDTNGQPQAL 3610 3620 3630 3640 3650 LVDGDQSKIL ISGLEPSTPY RFLLYGLHEG KRLGPLSAEG TTGLAPAGQT 3660 3670 3680 3690 3700 SEESRPRLSQ LSVTDVTTSS LRLNWEAPPG AFDSFLLRFG VPSPSTLEPH 3710 3720 3730 3740 3750 PRPLLQRELM VPGTRHSAVL RDLRSGTLYS LTLYGLRGPH KADSIQGTAR 3760 3770 3780 3790 3800 TLSPVLESPR DLQFSEIRET SAKVNWMPPP SRADSFKVSY QLADGGEPQS 3810 3820 3830 3840 3850 VQVDGQARTQ KLQGLIPGAR YEVTVVSVRG FEESEPLTGF LTTVPDGPTQ 3860 3870 3880 3890 3900 LRALNLTEGF AVLHWKPPQN PVDTYDVQVT APGAPPLQAE TPGSAVDYPL 3910 3920 3930 3940 3950 HDLVLHTNYT ATVRGLRGPN LTSPASITFT TGLEAPRDLE AKEVTPRTAL 3960 3970 3980 3990 4000 LTWTEPPVRP AGYLLSFHTP GGQNQEILLP GGITSHQLLG LFPSTSYNAR 4010 4020 4030 4040 4050 LQAMWGQSLL PPVSTSFTTG GLRIPFPRDC GEEMQNGAGA SRTSTIFLNG 4060 4070 4080 4090 4100 NRERPLNVFC DMETDGGGWL VFQRRMDGQT DFWRDWEDYA HGFGNISGEF 4110 4120 4130 4140 4150 WLGNEALHSL TQAGDYSMRV DLRAGDEAVF AQYDSFHVDS AAEYYRLHLE 4160 4170 4180 4190 4200 GYHGTAGDSM SYHSGSVFSA RDRDPNSLLI SCAVSYRGAW WYRNCHYANL 4210 4220 4230 4240 NGLYGSTVDH QGVSWYHWKG FEFSVPFTEM KLRPRNFRSP AGGG
INTENDED USE
The kit is a sandwich enzyme immunoassay for the in vitro quantitative measurement of TNX in human serum, plasma, tissue homogenates or other biological fluids.
DETECTION RANGE
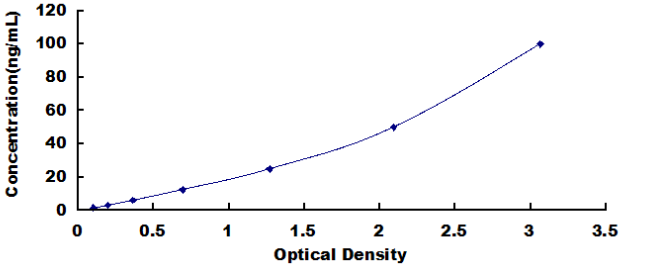
1.56-100ng/mL. The standard curve concentrations used for the ELISA’s were 100ng/mL, 50ng/mL, 25ng/mL, 12.5ng/mL, 6.25ng/mL, 3.12ng/mL, 1.56ng/mL.
SENSITIVITY
The minimum detectable dose of TNX is typically less than 0.53ng/mL.
The sensitivity of this assay, or Lower Limit of Detection (LLD) was defined as the lowest protein concentration that could be differentiated from zero. It was determined by adding two standard deviations to the mean optical density value of twenty zero standard replicates and calculating the corresponding concentration.
SPECIFICITY
This assay has high sensitivity and excellent specificity for detection of TNX.
No significant cross-reactivity or interference between TNX and analogues was observed.
You can reference link of the kit as following
https://dldevelop.com/Research-reagent/dl-tnx-hu.html
https://www.dldevelop.com/uploadfile/data/DL-TNX-Hu.pdf
Introduction
| Item | Standard | Test | |
| Description |
The kit is a sandwich enzyme immunoassay for the in vitro quantitative measurement of TNX in human serum, plasma, tissue homogenates and other biological fluids. |
Conform | |
| Identification | Colorimetric | Positive | |
| Composition | Traditional ELISA Kit | Ready-to-Use ELISA KIT | Conform |
| Pre-coated, ready to use 96-well strip plate 1 | Pre-coated, ready to use 96-well strip plate 1 | ||
| Plate sealer for 96 wells 2 | Plate sealer for 96 wells 2 | ||
| Standard 2 | Standard 2 | ||
| Diluents buffer 1×45mL | Standard Diluent 1×20mL | ||
| Detection Reagent A 1×120μL | Detection Solution A 1×12mL | ||
| Detection Reagent B 1×120μL | Detection Solution B 1×12mL | ||
| TMB Substrate 1×9mL | TMB Substrate 1×9mL | ||
| Stop Solution 1×6mL | Stop Solution 1×6mL | ||
| Wash Buffer (30 × concentrate) 1×20mL | Wash Buffer (30 × concentrate) 1×20mL | ||
| Instruction manual 1 | Instruction manual 1 | ||
Test principle
The microtiter plate provided in this kit has been pre-coated with an antibody specific to the index. Standards or samples are then added to the appropriate microtiter plate wells with a biotin-conjugated antibody preparation specific to the index. Next, Avidin conjugated to Horseradish Peroxidase (HRP) is added to each microplate well and incubated. After TMB substrate solution is added, only those wells that contain the index, biotin-conjugated antibody and enzyme-conjugated Avidin will exhibit a change in color. The enzyme-substrate reaction is terminated by the addition of sulphuric acid solution and the color change is measured spectrophotometrically at a wavelength of 450nm ± 10nm. The concentration of the index in the samples is then determined by comparing the O.D. of the samples to the standard curve.
Recovery
Matrices listed below were spiked with certain level of recombinant TNX and the recovery rates were calculated by comparing the measured value to the expected amount of the index in samples.
| Matrix | Recovery range (%) | Average(%) |
| serum(n=5) | 81-93 | 86 |
| EDTA plasma(n=5) | 80-97 | 88 |
| heparin plasma(n=5) | 90-101 | 95 |
Linearity
The linearity of the kit was assayed by testing samples spiked with appropriate concentration of the index and their serial dilutions. The results were demonstrated by the percentage of calculated concentration to the expected.
| Sample | 1:2 | 1:4 | 1:8 | 1:16 |
| serum(n=5) | 82-96% | 83-98% | 81-99% | 93-101% |
| EDTA plasma(n=5) | 88-101% | 86-95% | 90-102% | 80-93% |
| heparin plasma(n=5) | 80-91% | 82-90% | 95-104% | 79-95% |
Precision
Intra-assay Precision (Precision within an assay): 3 samples with low, middle and high level the index were tested 20 times on one plate, respectively.
Inter-assay Precision (Precision between assays): 3 samples with low, middle and high level the index were tested on 3 different plates, 8 replicates in each plate.
CV(%) = SD/meanX100
Intra-Assay: CV<10%
Inter-Assay: CV<12%
Stability
The stability of ELISA kit is determined by the loss rate of activity. The loss rate of this kit is less than 5% within the expiration date under appropriate storage conditions.
Note:
To minimize unnecessary influences on the performance, operation procedures and lab conditions, especially room temperature, air humidity and incubator temperatures should be strictly regulated. It is also strongly suggested that the whole assay is performed by the same experimenter from the beginning to the end.
Assay procedure summary
1. Prepare all reagents, samples and standards;
2. Add 100µL standard or sample to each well. Incubate 2 hours at 37℃;
3. Aspirate and add 100µL prepared Detection Reagent A. Incubate 1 hour at 37℃;
4. Aspirate and wash 3 times;
5. Add 100µL prepared Detection Reagent B. Incubate 1 hour at 37℃;
6. Aspirate and wash 5 times;
7. Add 90µL Substrate Solution. Incubate 15-25 minutes at 37℃;
8. Add 50µL Stop Solution. Read at 450nm immediately.
Order or get a Quote
We will reply you within 24 hours!














